Host (772545)
Bovine (1090)Canine (20)Cat (403)Chicken (1642)Cod (2)Cow (333)Crab (15)Dog (524)Dolphin (2)Duck (13)E Coli (239120)Equine (7)Feline (1864)Ferret (306)Fish (125)Frog (52)Goat (37446)Guinea Pig (752)Hamster (1376)Horse (903)Insect (2053)Mammalian (512)Mice (6)Monkey (624)Mouse (96940)Pig (197)Porcine (70)Rabbit (361350)Rat (11751)Ray (55)Salamander (4)Salmon (15)Shark (3)Sheep (4248)Snake (4)Swine (301)Turkey (57)Whale (3)Yeast (5335)Zebrafish (3022)Isotype (158144)
IgA (13516)IgA1 (919)IgA2 (319)IgD (1995)IgE (5499)IgG (88235)IgG1 (16852)IgG2 (1331)IgG3 (2728)IgG4 (1696)IgM (22319)IgY (2735)Label (241822)
AF488 (2868)AF594 (719)AF647 (2711)ALEXA (12104)ALEXA FLUOR 350 (298)ALEXA FLUOR 405 (303)ALEXA FLUOR 488 (715)ALEXA FLUOR 532 (303)ALEXA FLUOR 555 (317)ALEXA FLUOR 568 (296)ALEXA FLUOR 594 (342)ALEXA FLUOR 633 (305)ALEXA FLUOR 647 (650)ALEXA FLUOR 660 (295)ALEXA FLUOR 680 (465)ALEXA FLUOR 700 (2)ALEXA FLUOR 750 (457)ALEXA FLUOR 790 (258)Alkaline Phosphatase (825)Allophycocyanin (32)ALP (387)AMCA (133)AP (1213)APC (14722)APC C750 (13)Apc Cy7 (1248)ATTO 390 (3)ATTO 488 (6)ATTO 550 (1)ATTO 594 (5)ATTO 647N (4)AVI (53)Beads (235)Beta Gal (2)BgG (1)BIMA (6)Biotin (27912)Biotinylated (1810)Blue (708)BSA (878)BTG (46)C Terminal (688)CF Blue (19)Colloidal (22)Conjugated (29545)Cy (163)Cy3 (445)Cy5 (2092)Cy5 5 (2520)Cy5 PE (1)Cy7 (3689)Dual (170)DY549 (3)DY649 (3)Dye (1)DyLight (1459)DyLight 405 (11)DyLight 488 (223)DyLight 549 (22)DyLight 594 (90)DyLight 649 (4)DyLight 650 (35)DyLight 680 (20)DyLight 800 (24)Fam (13)Fc Tag (8)FITC (29672)Flag (223)Fluorescent (146)GFP (576)GFP Tag (180)Glucose Oxidase (59)Gold (511)Green (580)GST (722)GST Tag (327)HA Tag (455)His (649)His Tag (522)Horseradish (550)HRP (13090)HSA (247)iFluor (16571)Isoform b (31)KLH (87)Luciferase (105)Magnetic (260)MBP (343)MBP Tag (93)Myc Tag (425)OC 515 (1)Orange (78)OVA (103)Pacific Blue (213)Particle (64)PE (34110)PerCP (7934)Peroxidase (1364)POD (11)Poly Hrp (94)Poly Hrp40 (13)Poly Hrp80 (3)Puro (32)Red (2491)RFP Tag (63)Rhodamine (612)RPE (910)S Tag (194)SCF (184)SPRD (351)Streptavidin (55)SureLight (77)T7 Tag (97)Tag (4866)Texas (1300)Texas Red (1282)Triple (10)TRITC (1457)TRX tag (90)Unconjugated (2116)Unlabeled (218)Yellow (84)Pathogen (489310)
Adenovirus (8679)AIV (317)Bordetella (25032)Borrelia (18284)Candida (17799)Chikungunya (640)Chlamydia (17579)CMV (121402)Coronavirus (5949)Coxsackie (863)Dengue (2875)EBV (1485)Echovirus (215)Enterovirus (677)Hantavirus (259)HAV (909)HBV (2084)HHV (838)HIV (7877)hMPV (275)HSV (2335)HTLV (635)Influenza (22006)Isolate (1208)KSHV (396)Lentivirus (3755)Lineage (2945)Lysate (127759)Marek (94)Measles (1169)Parainfluenza (1692)Poliovirus (3031)Poxvirus (81)Rabies (1526)Reovirus (536)Retrovirus (1066)Rhinovirus (511)Rotavirus (5354)RSV (1768)Rubella (1070)SIV (279)Strain (67791)Vaccinia (7234)VZV (661)WNV (370)Species (2892355)
Alligator (10)Bovine (151363)Canine (112144)Cat (13127)Chicken (105456)Cod (1)Cow (2031)Dog (12763)Dolphin (21)Duck (9734)Equine (2088)Feline (1019)Ferret (259)Fish (12886)Frog (1)Goat (81773)Guinea Pig (79203)Hamster (36739)Horse (41326)Human (949911)Insect (653)Lemur (121)Lizard (24)Monkey (102322)Mouse (466874)Pig (26375)Porcine (123452)Rabbit (119266)Rat (342786)Ray (451)Salmon (350)Seal (8)Shark (29)Sheep (96357)Snake (11)Swine (519)Toad (4)Turkey (244)Turtle (75)Whale (45)Zebrafish (534)Technique (11605397)
Activation (211947)Activity (15345)Affinity (89577)Agarose (3204)Aggregation (229)Antigen (172177)Apoptosis (28670)Array (3334)Blocking (105047)Blood (10636)Blot (15167)ChiP (1048)Chromatin (6185)Colorimetric (14483)Control (90316)Culture (5592)Cytometry (5669)Depletion (122)DNA (183052)Dot (315)EIA (1082)Electron (6383)Electrophoresis (438)ELISA (2989935)Elispot (2324)Enzymes (56005)Exosome (4927)Extract (1465)Fab (3119)FACS (61)FC (98276)Filter Tips (388)Flow (14262)Fluorometric (1559)Formalin (122)Frozen (22754)Functional (730)Gel (3362)HTS (174)IF (12322)IHC (27118)Immunoassay (8462)Immunofluorescence (5625)Immunohistochemistry (81)Immunoprecipitation (80)intracellular (5792)IP (4381)iPSC (342)Isotype (17654)Lateral (1806)Lenti (382497)Light (44969)Microarray (203)MicroRNA (4886)Microscopy (808)miRNA (134352)Monoclonal (1477760)Multi (5178)Multiplex (7702)Negative (4545)PAGE (2639)Panel (2360)Paraffin (2732)PBS (27864)PCR (9)Peptide (336176)PerCP (67522)Polyclonal (3789572)Positive (6593)Precipitation (84)Premix (155)Primers (47311)Probe (3175)Profile (561)Pure (10551)Purification (15)Purified (111252)Real Time (3379)Resin (3256)Reverse (3119)RIA (521)RNAi (63116)Rox (1339)RT PCR (6831)Sample (4733)SDS (1885)Section (2971)Separation (246)Sequencing (222)Shift (21)siRNA (587499)Standard (74021)Sterile (11995)Strip (2479)Taq (2)Tip (1721)Tissue (83609)Tube (4395)Vitro (3588)Vivo (2366)WB (3025)Western Blot (14443)Tissue (2006372)
Adenocarcinoma (1075)Adipose (3400)Adrenal (645)Adult (4885)Amniotic (65)Animal (2451)Aorta (436)Appendix (89)Array (2022)Ascites (4737)Bile Duct (20)Bladder (1639)Blood (8502)Bone (27167)Brain (30948)Breast (10816)Calvaria (28)Carcinoma (13377)cDNA (58549)Cell (412333)Cellular (9299)Cerebellum (700)Cervix (232)Child (1)Choroid (19)Colon (3865)Connective (3593)Contaminant (3)Control (80438)Cord (661)Corpus (148)Cortex (686)Dendritic (1827)Diseased (265)Donor (1360)Duct (831)Duodenum (643)Embryo (425)Embryonic (4560)Endometrium (454)Endothelium (1426)Epidermis (166)Epithelium (4208)Esophagus (716)Exosome (4282)Eye (1951)Female (475)Frozen (2671)Gallbladder (155)Genital (5)Gland (3433)Granulocyte (8920)Heart (6825)Hela (413)Hippocampus (325)Histiocytic (74)Ileum (201)Insect (4880)Intestine (1945)Isolate (1208)Jejunum (175)Kidney (8052)Langerhans (283)Leukemia (21444)Liver (17188)Lobe (823)Lung (6026)Lymph (1208)Lymphatic (639)lymphocyte (22427)Lymphoma (12623)Lysate (127759)Lysosome (2778)Macrophage (31742)Male (1605)Malignant (1457)Mammary (1965)Mantle (1042)Marrow (2190)Mastocytoma (3)Matched (11710)Medulla (156)Melanoma (15385)Membrane (105110)Metastatic (3581)Mitochondrial (156958)Muscle (37181)Myeloma (748)Myocardium (11)Nerve (6403)Neuronal (16836)Node (1206)Normal (9490)Omentum (10)Ovarian (2477)Ovary (1161)Pair (47185)Pancreas (2822)Panel (1633)Penis (64)Peripheral (1918)Pharynx (122)Pituitary (5346)Placenta (4037)Prostate (9376)Proximal (318)Rectum (316)Region (201823)Retina (956)Salivary (3096)Sarcoma (6944)Section (2895)Serum (24827)Set (167584)Skeletal (13569)Skin (1860)Smooth (7529)Spinal (424)Spleen (2276)Stem (8873)Stomach (922)Stroma (49)Subcutaneous (47)Testis (15243)Thalamus (127)Thoracic (60)Throat (42)Thymus (2974)Thyroid (13931)Tongue (144)Total (9948)Trachea (227)Transformed (175)Tubule (48)Tumor (76618)Umbilical (208)Ureter (73)Urinary (2418)Robust Bacterial scRNA-seq Protocol Developed: A New Chapter in Microbial Transcriptomics
Gentaur
Scientific Publications
Robust Bacterial scRNA-seq Protocol Developed: A New Chapter in Microbial Transcriptomics
Introduction
In the expanding field of single-cell technologies, the application of single-cell RNA sequencing (scRNA-seq) to bacterial systems has long been a technical frontier. Unlike their eukaryotic counterparts, bacterial cells present a complex set of barriers for single-cell transcriptomic profiling, including the absence of polyadenylated RNA, extremely low mRNA abundance, and a robust cell wall structure that resists lysis. Today, the development of a robust bacterial scRNA-seq protocol represents a significant methodological advancement, enabling high-throughput, high-resolution analysis of bacterial gene expression at the single-cell level.
This protocol paves the way for analyzing population heterogeneity, phenotypic variation, and stress responses in microbiology research. In this blog post, we’ll explore the technical background, methodological breakthroughs, and practical implications of this protocol for scientific research, biotechnology applications, and academic investigations.
What is Single-Cell RNA Sequencing in Bacteria?
Single-cell RNA sequencing (scRNA-seq) is a tool used to measure gene expression in individual cells. While commonly used in mammalian and yeast systems, its implementation in bacteria was until recently limited due to the technical difficulty of isolating and amplifying bacterial mRNA from single cells.
Traditional bulk RNA-seq averages gene expression across a population, often masking rare subpopulations or transient states. scRNA-seq resolves this by analyzing each cell independently. In bacterial populations, such granularity is critical for understanding cell-to-cell variation in processes like antibiotic response, sporulation, biofilm formation, and metabolic adaptation.
To date, several papers including those from ncbi.nlm.nih.gov, genome.gov, and nih.gov have outlined early attempts at adapting eukaryotic scRNA-seq platforms for bacteria. However, many of these early adaptations suffered from low capture efficiency, high technical noise, and lack of scalability.
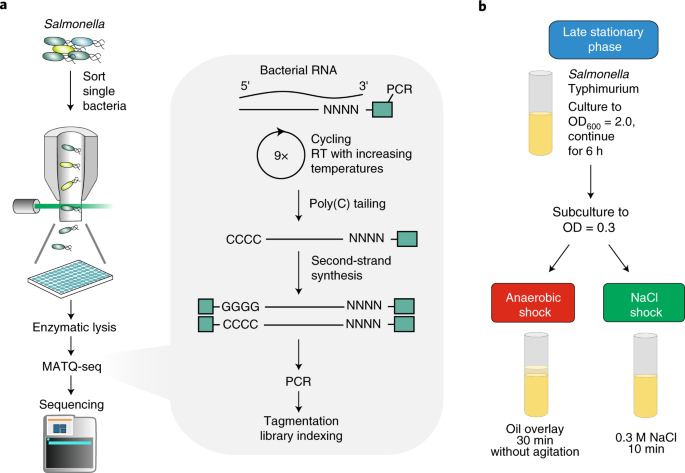
Core Challenges in Bacterial scRNA-seq
Bacterial cells pose unique technical challenges:
- Low mRNA Abundance: Bacterial cells typically contain 100 to 1,000 mRNA molecules at any given time, compared to tens of thousands in a typical mammalian cell.
- Lack of Poly(A) Tails: Most scRNA-seq methods rely on poly(A) capture. Bacterial RNAs do not naturally possess these tails.
- Tough Cell Wall: Gram-positive and Gram-negative bacteria have rigid peptidoglycan layers that complicate lysis and RNA extraction.
- Fast RNA Turnover: Bacterial transcripts degrade quickly, requiring rapid and efficient preservation.
These challenges necessitate novel approaches that are distinct from standard mammalian protocols.
The Breakthrough: A New Robust Protocol
The newly developed bacterial scRNA-seq protocol addresses these issues using several integrated strategies:
1. Microfluidic Isolation of Single Bacterial Cells
Leveraging modified microfluidic devices originally used in Drop-seq and 10x Genomics platforms, researchers adapted the system for bacterial size and rigidity. Cell encapsulation in droplets ensures physical isolation, minimizing RNA contamination between cells.
2. In Situ Cell Wall Disruption
The protocol uses a cocktail of enzymatic treatments (e.g., lysozyme, proteinase K) in combination with mechanical disruption to achieve efficient lysis without compromising RNA integrity. Optimization of osmotic buffers ensures minimal stress-induced gene expression artifacts during cell preparation.
3. Tailored RNA Capture System
To address the lack of poly(A) tails, a synthetic poly(A) tailing step using E. coli poly(A) polymerase is introduced post-lysis. This enables capture by oligo-dT primers during reverse transcription.
4. RNA Amplification and UMIs
To combat low mRNA copy number, a modified Smart-seq2 approach is used with early incorporation of unique molecular identifiers (UMIs) to minimize amplification bias and improve quantification.
5. RNA Stabilization Techniques
The protocol incorporates RNA stabilization agents such as RNAlater, combined with flash freezing, to preserve transcripts during processing.
Detailed parameters and enzyme ratios are available in publications listed on PubMed and protocols archived on protocols.io.
Applications in Microbiology Research
This protocol opens up new opportunities for research in microbiology:
🔬 Understanding Population Heterogeneity
Bacterial populations often consist of diverse subpopulations that behave differently even in clonal cultures. With this method, researchers can now identify slow-growing persisters, stress-resistant cells, and metabolic outliers.
🧪 Monitoring Stress Responses
Researchers can monitor the gene expression profile of individual cells exposed to osmotic stress, oxidative environments, or nutrient deprivation—essential for understanding adaptation mechanisms.
🧫 Biofilm and Community Studies
By combining this protocol with spatial transcriptomics, it's possible to study microbial communities, including biofilms and microbiomes, with cell-type and location resolution.
🧭 Synthetic Biology and Engineering
The ability to monitor synthetic circuits in individual bacteria allows for real-time evaluation of promoter strength, noise, and expression heterogeneity in engineered strains.
For more technical insights, see datasets shared on Gene Expression Omnibus and visualization tools provided by UCSC Genome Browser.

Future Developments and Optimizations
Although this protocol represents a major advancement, several areas remain under active development:
- Improved mRNA capture efficiency to reduce dropout rates
- Automated cell lysis-on-chip to scale up throughput
- Barcoding innovations to multiplex hundreds of samples
- Multi-omics integration with proteomics and metabolomics workflows
- Standardization across bacterial species, including extremophiles
Final Thoughts: A Milestone for Bacterial Transcriptomics
The development of a robust bacterial scRNA-seq protocol represents a transformative step in microbiological research. It allows scientists to go beyond bulk averages and instead focus on the richness of single-cell diversity. From uncovering hidden bacterial states to tracking gene expression noise in synthetic circuits, the applications are vast and expanding.
This tool is now essential for labs focused on microbial physiology, pathogenesis, environmental microbiology, synthetic biology, and systems biology. The future of bacterial research will be increasingly single-cell–driven, and this protocol provides the foundation for that shift.
Tags
- Biology
- LifeSciences
- gentaur
- Biodiversity
- Evolution
- Genetics
- MolecularBiology
- CellBiology
- DNA
- Biotechnology
- Research
